CRS4 was a pioneer in the use of the MapReduce programming paradigm for the scalable processing of sequencing data.
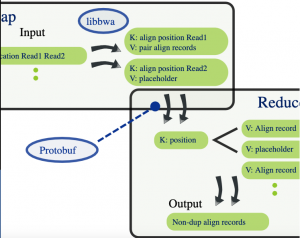
Among its various contributions in this context, CRS4 produced one of the early programs that used this paradigm to compute the DNA sequence mapping and identify duplicate sequences due to the biological sample preparation process.
To help integrate this sophisticated technology into established laboratory practices and make it more accessible to end users, CRS4 integrated these components into a widely used web-based workflow management system effectively assembling a simple-to-use data analysis platform backed by the scalability and computing power of CRS4’s tools and computing infrastructure.
The following article introduced SEAL, one of the first Hadoop-based DNA sequencing alignment programs along with an accompanying toolkit for distributed DNA sequence processing. It was the first in a series of CRS4 contributions aimed at providing scalable sequence processing on commodity hardware, instead of relying on expensive large computing nodes and large cluster-shared file systems.
- L Pireddu, S Leo, G Zanetti. SEAL: a distributed short read mapping and duplicate removal tool Bioinformatics 27 (15), 2011: 2159-2160.