Available standards and guidelines, and how CRS4’s activities contribute to them
In this focus Alessandro Sulis (Healthcare Flows group, CRS4) will discuss the need for interoperability between clinical systems, the available standards and guidelines, and how CRS4’s activities contribute to them.
In modern hospitals it is crucial that different clinical domains (e.g., laboratory, radiology, etc.) be able to exchange information. From the technical point of view, this requirement implies that different domains need to be able to exchange mutually understandable messages carrying information about patients and healthcare processes. This apparently simple requirement is difficult to satisfy in practice. In fact, clinical systems tend to be designed as separate “information islands” that do not easily communicate with the rest of the world.
As a reaction, clinical informatics interoperability standards and guidelines have been developed in the past twenty years and are now starting to be widely supported by vendors. CRS4 is actively contributing to the development of these standards and guidelines – particularly the ones in the Pathology and Laboratory domains – and to their implementation in real-world clinical systems.

Alessandro Sulis, Graduated in Electronic Engineering at the University of Cagliari and II level Master in Information Technology, at the Center of Excellence CEFRIEL in Milan. He works at CRS4 since 2007, as a technologist involved on different research projects about biomedical and clinical engineering applied to real clinical contexts. Currently he is working on clinical systems interoperability, following the main standards and best practices; he strictly collaborates with the reference organizations (IHE and HL7 international): in particular, he is a member of the IHE PaLM (Pathology and Laboratory Medicine) domain Technical Committee.
(with english subtitles gently generated by an algorithm by Gavin Brelstaff)
— E. Falqui
Technical Corner
Health IT interoperability has an important role in health service quality and effectiveness at different levels, from the intra-institutional communication between department-based clinical systems (such as Laboratory Information Systems, Radiology Information Systems, etc.) to the inter-institutional interaction and coordination of regional or national components of a healthcare system. Also, in clinical and biomedical research, data integration is essential as a basis for guiding differential outcome analysis. Interoperability implies an exchange and mutual interpretation of information by two or more systems. In fact, the first definition of interoperability in the IEEE Standard Computer Dictionary stated, “the ability of two or more systems or components to exchange information and to use the information that has been exchanged”1. Notice that the concept is not limited to information exchange, since this action is necessary but not sufficient to achieve interoperability. Focusing on health informatics, HL7 International2 gave a more detailed characterization3 identifying three main types of health IT interoperability:
- technical interoperability, focusing on physical communication of data;
- semantic interoperability, relating to the meaning of data exchanged;
- process interoperability, addressing systems integration into a work setting.
This classification goes beyond simple labels, creating a hierarchical model which can guide interoperability implementation in different contexts: the activity of Healthcare Flows Research Program spans all the three types above and the following paragraphs show some of the results obtained in this field.
Technical Interoperability
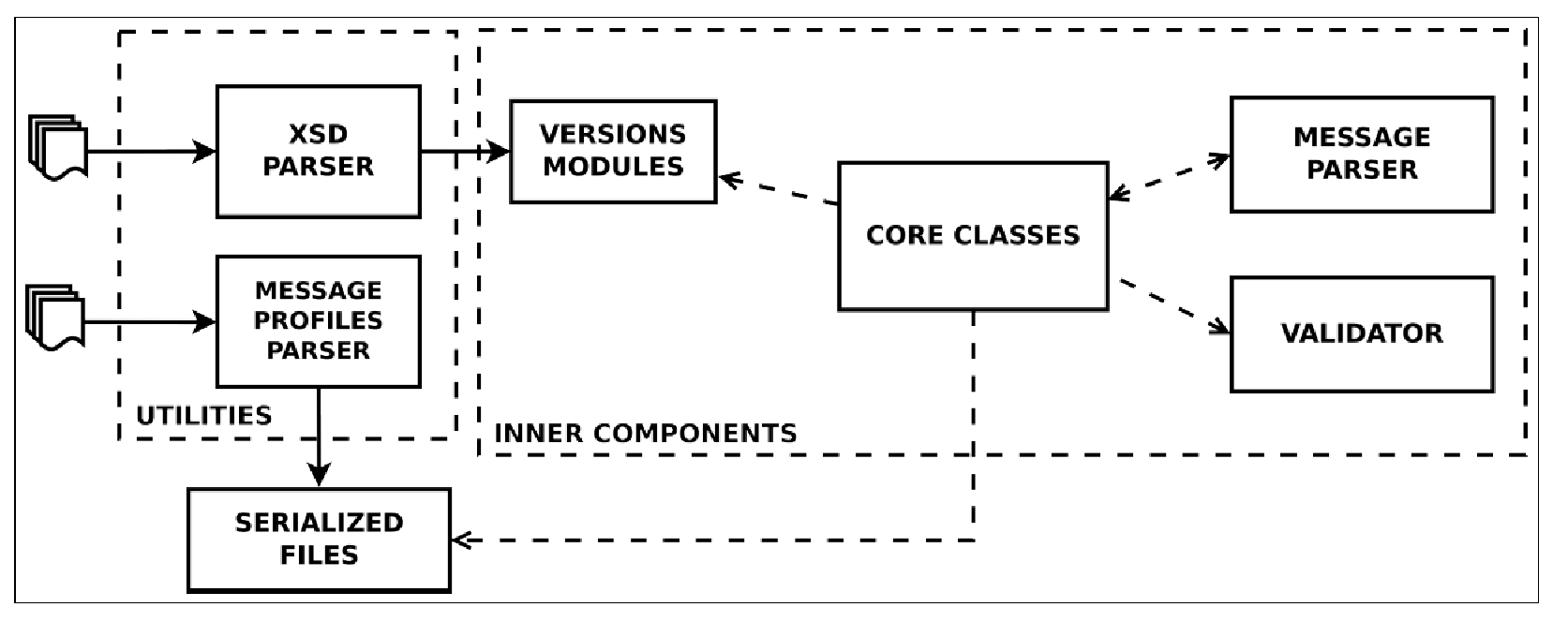
From the technical interoperability perspective, the group develops and maintains HL7apy, an innovative Python package to manage HL7 v2.x messages for fast prototyping of HL7-compliant software. As shown in Figure 1, the main library components are:
- inner components: core classes and an API to create and manage HL7 messages; Message Parser, to parse ER7-encoded messages; and Validator, to validate messages;
- two utility scripts: an XSD Parser, to generate Python modules for every HL7 v2 minor version; and a Message Profiles Parser, to serialize files for message profiles.
The main classes defined in HL7apy to represent HL7 elements are:
- Messages
- Group
- Segment
- Field
- Component
- SubComponent
- Base datatype classes
The core classes allow the developer to create HL7-compliant messages, to navigate their structure and to manipulate HL7 elements in a compact form, as shown in the code examples in Figure 2, Figure 3 and Figure 4.
One of the most important features provided by HL7apy is the validation of messages, since it ensures their compliance with the standard for the specific message type and HL7 version. The library allows two levels of validation: STRICT and TOLERANT.
A detailed description of HL7apy can be found in4; the project is still under development and the code is released under an open-source license5.
Semantic and Process interoperability
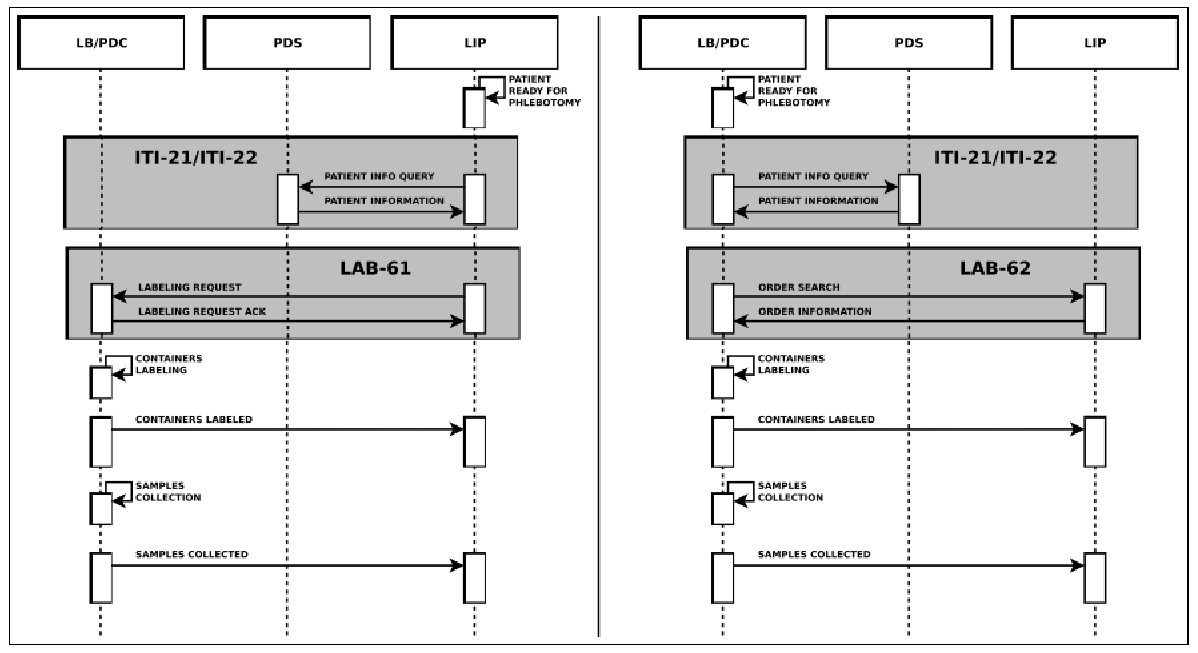
The Healthcare Flows group also actively works on semantic and process interoperability, collaborating intensely with IHE6– the reference institution for information sharing and healthcare systems interoperability. In 2010 the group joined the IHE Technical Committee for the Laboratory Domain, now called “Pathology and Laboratory Medicine (PaLM)” Domain. Our first contribution in this context was the proposal of an extension to the IHE Laboratory guidelines to enhance traceability in the pre-analytical phase, going from the prescription of exams to sample preparation for analysis7. For each domain, IHE models clinical processes by defining the main workflow steps in detail, the main use cases, the actors involved, the transactions and, for each transaction, the messages to be used and the specific meaning of each message field. Concerning the laboratory pre-analytical process, IHE defines two main steps, identification and sample collection, mapping them to IHE PDQ (Patient Demographics Query) and LBL (Laboratory Specimen Barcode Labeling) profiles. The actors involved are:
- Patient Demographics Consumer (PDC): to query a set of patient information;
- Patient Demographic Supplier (PDS): provides patient-specific information;
- Label information Provider (LIP): the actor that provides information about labels;
- Label Broker (LB): the actor responsible for the labeling of the containers according to the information provided by the LIP.
The transactions for the two profiles are:
- ITI-21 and ITI-22 for PDQ;
- LAB-61 and LAB-62 for LBL
All of them are based on HL7 messages exchange. Concerning LBL, in a phlebotomy process supported by automation, the robotized labeling of specimen containers considers two different scenarios: Request Mode and Query Mode. These are both showed in Figure 5.
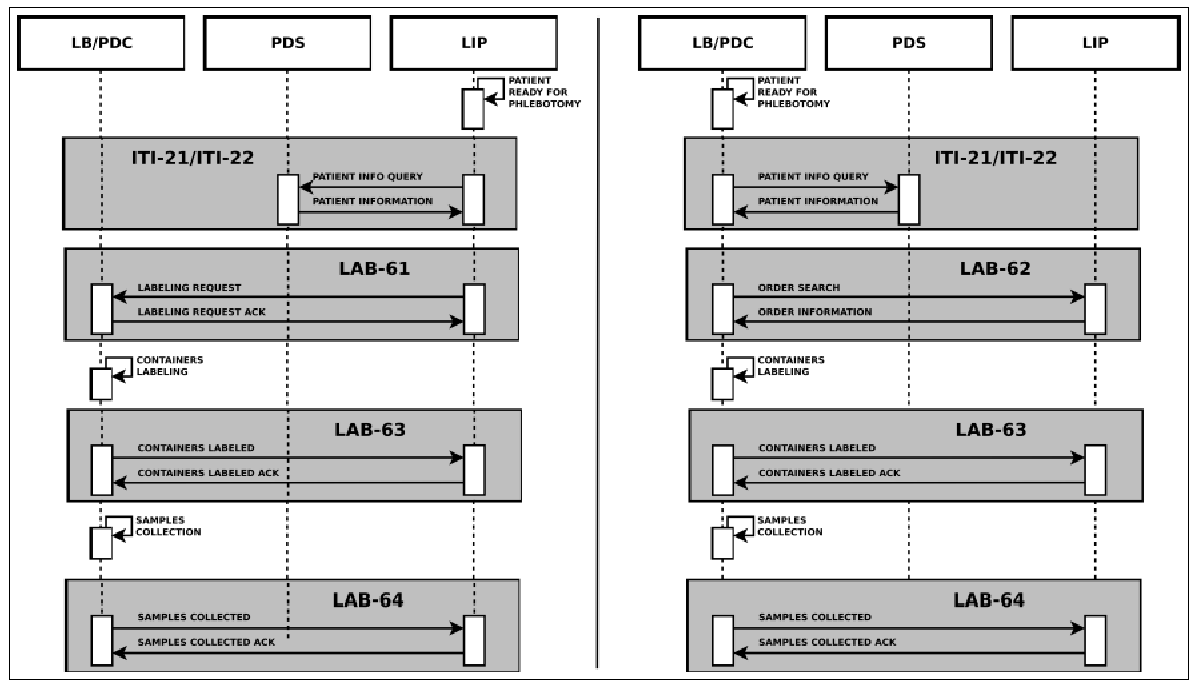
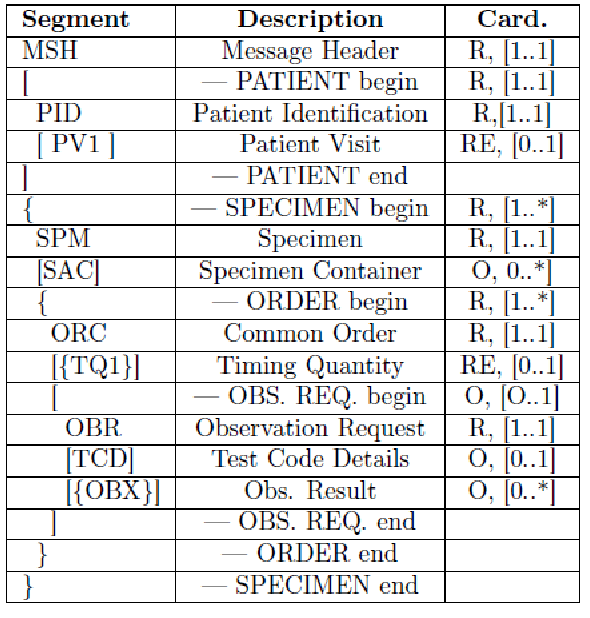
The main information needed for the labeling process is carried by the HL7 messages exchanged between the actors. In detail they provide: patient data, drawn specimens with their unique id, tests to be performed on every specimen and type of container to use. This information is very useful for traceability purposes, but no details are provided by the LBL profile regarding the preparation of the sample container and the sample collection. For these reasons, we proposed an extension of this profile, adding two new transactions, LAB-63 (Labeled Containers Production Confirmation) and LAB-64 (Specimen Collection Confirmation), depicted in Figure 6.
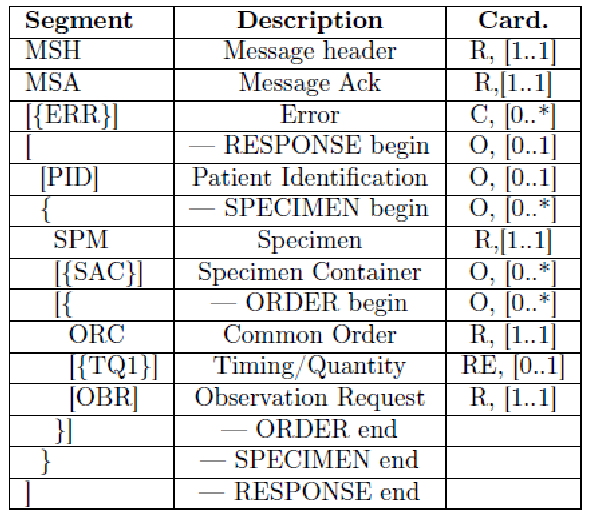
The HL7 messages chosen to define these transactions are OML^O33 (Laboratory Order Message) and ORL^O34 for acknowledgment, since they are specimen-centric and their structures contain all the information needed for traceability purposes.
We submitted the profile extension in a Supplement to the Committee for public discussion in July 2011. LAB-63 was reviewed and immediately accepted and – after the trial implementation period – was included in IHE PaLM Domain Technical Framework8. LAB-64 was considered above LBL scope, as it also involves other IHE profiles; for these reasons, the proposed profile became the starting point for a completely new profile, called Specimen Event Tracking (SET), which we have been developing since 2016 in strict collaboration with PALM Technical Committee.
http://www.en13606.org/the-ceniso-en13606-standard/semantic-interoperability ↩
HL7 EHR Interoperability Work Group, “Coming to terms”, available at https://www.hln.com/assets/pdf/Coming-to-Terms-February-2007.pdf ↩
Meloni V. et al, “HL7apy: a Python library to parse, create and handle HL7 v2.x messages” ”, EJBI - European Journal for Biomedical Informatics, Volume: 12, 2016, available at http://www.ejbi.org/scholarly-articles/hl7apy-a-python-library-to-parse-create-and-handle-hl7v2x-messages.pdf ↩
Sulis A. et al, “Traceability Based Description of Clinical Processes: Extension of IHE Guidelines for Phlebotomy Workflows”, EJBI - European Journal for Biomedical Informatics, Volume: 12, 2016, available at http://ihic2016.eu/Doc/02_2016_06_13_HL7apy@IHIC_Sulis.pdf ↩